


BlastoSPIM web site main entry page.

|

|
| Ground truth segmentation of nuclei in late blastocyst embryo. | System segmentation of nuclei in same late blastocyst embryo. |
|---|
We introduce new ground truth datasets of 3D nuclear instance segmentation in pre-implantation mouse embryos as well as models trained on those datasets. One ground truth dataset (BlastoSPIM 1.0) is primarily composed of embryos from the 8-nuclei stage to the 64-nuclei stage. The other ground truth dataset (BlastoSPIM 2.0) is primarily composed of embryos from the 64-nuclei stage to the >100-nuclei stage. BlastoSPIM 1.0 and 2.0 are two of the largest and most complete datasets of their kind with 573 and 80 annotated light-sheet images, respectively. Each image has an xy-resolution of 0.208 micron and a z-resolution of 2.0 microns. For each of the 18336 nuclei across the two datasets, the ground-truth instance is defined by drawing a contour in each z-slice in which the nucleus appears.
Using this dataset, we have built a model to accurately perform 3D instance segmentation and begun several investigations into the quantitative analysis of embryogenesis including fate specification, nuclear volume and shape variations, and embryo viability. This model is also the first step in lineage construction and opens the potential for a more direct exploration into early development.
Significant progress has been made in providing a means to segment nuclei in 2D [Cellpose 2.0] but there are very few biological datasets with complete ground truth instance segmentation of nuclei in 3D. This dataset can be used to help enable transfer learning for other applications via pre-training, fine-tuning, simulating, augmenting, evaluating etc.
Hayden Nunley, Binglun Shao,Prateek Grover, Jaspreet Singh, Bradley Joyce, Rebecca Kim-Yip, Abraham Kohrman, Aaron Watters, Zsombor Gal, Alison Kickuth, Madeleine Chalifoux, Stanislav Shvartsman, Eszter Posfai, Lisa M. Brown
Center for Computational Biology, Flatiron Institute - Simons Foundation, New York, United States of America
Department of Chemical and Biological Engineering, Princeton University, Princeton, New Jersey, United States of America
Department of Molecular Biology, Princeton University, Princeton, New Jersey, United States of America
The Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, New Jersey, United States of America
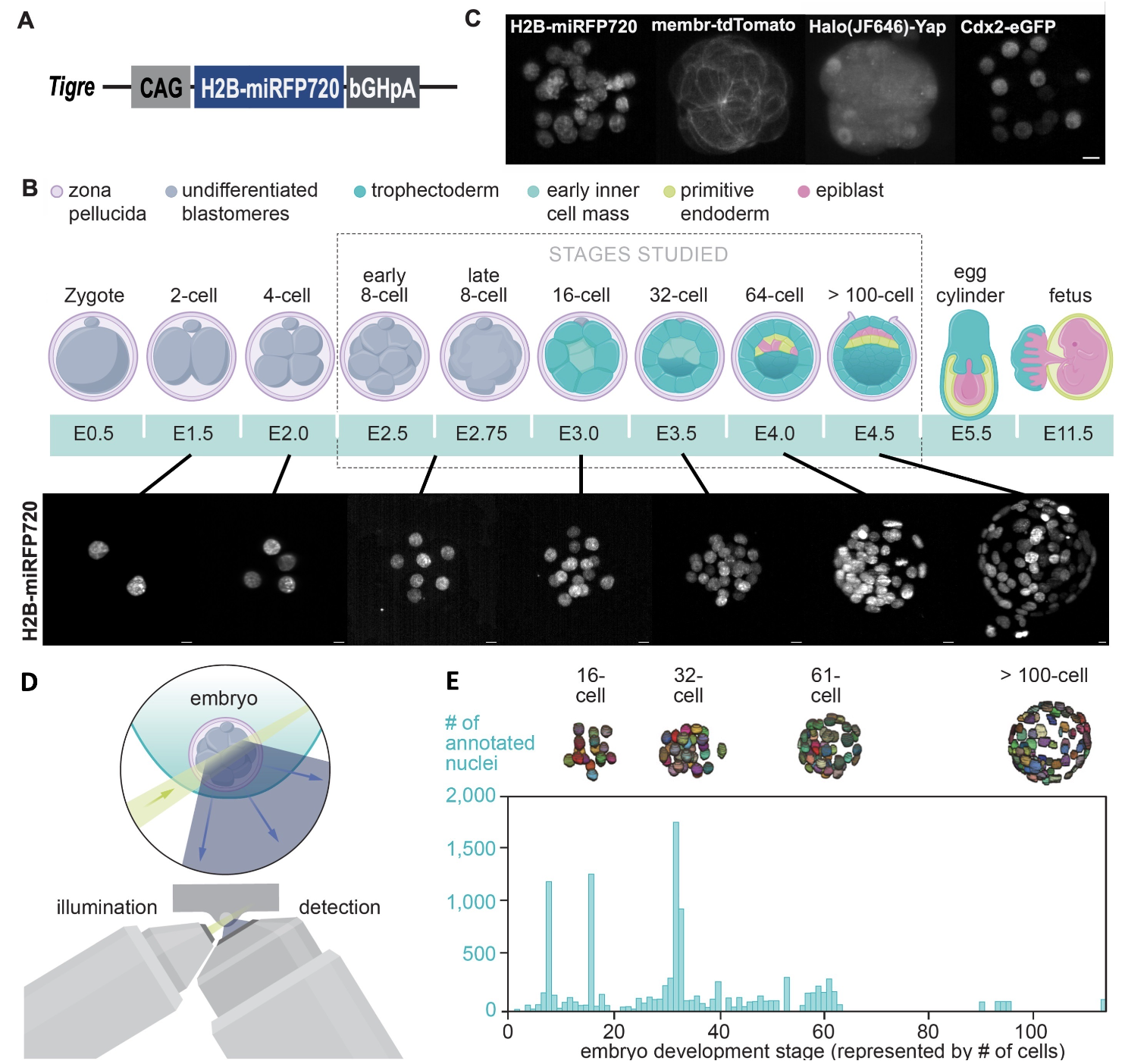
Paper Fig 3. BlastoSPIM, ground truth of nuclear instance segmentation for embryos expressing a novel near-infrared nuclear marker. (A) Schematic of targeted TIGRE locus with the CAG-H2B-miRFP720 insert. (B) Top: cartoon of preimplantation development in the mouse. After fertilization, the zygote undergoes rounds of division in the oviduct. At the 8-cell stage, compaction and polarization occur. By the 32-cell stage, a subset of cells called the trophectoderm (TE) form the embryo’s surface; the remaining cells form the inner cell mass (ICM). The ICM cells begins to pattern into two fates, primitive endoderm (PE) and epiblast (EPI), by the 64-cell stage; by implantation, at the > 100-cell stage, the two inner fates are spatially segregated. Bottom: Maximum-intensity projected images – acquired with SPIM – of preimplantation embryos expressing H2B-miRFP720 at different developmental stages. Scale bar: 10 μm. (C) Preimplantation embryo expressing four spectrally distinct fluorescent reporters: CDX2-eGFP; membrane-tdTomato (mT/mG); Halo-YAP and H2B-miRFP720. Maximum intensity projections of images acquired with SPIM. Scale bar: 10 μm. (D) Live imaging preimplantation development. Green: light sheet used for illumination. Blue: emitted light is collected by the detection objective. (E) Histogram of number of nuclei per embryonic stage (represented by embryo cell number). For four example embryos from different stages, the ground truth of nuclear segmentation are shown.

Time lapse sequence of the nuclei in the early mouse embryo as acquired using a light-sheet microscope every 15 minutes, shown in maximum intensity projection.